ChemMORT
ChemMORT (Chemical Molecular Optimization, Representation and Translation) is a platform for de novo molecular optimization by navigating chemical space based on neural machine translation and inverse QSAR methodology. Based on reversible molecular representation and multi-objective particle swarm optimization, ChemMORT could effectively optimize any given molecule toward some predefined ADMET endpoints with the preservation of potency by integrating the pre-trained ADMET prediction models, substructural constraint and similarity constraint.
Calculation
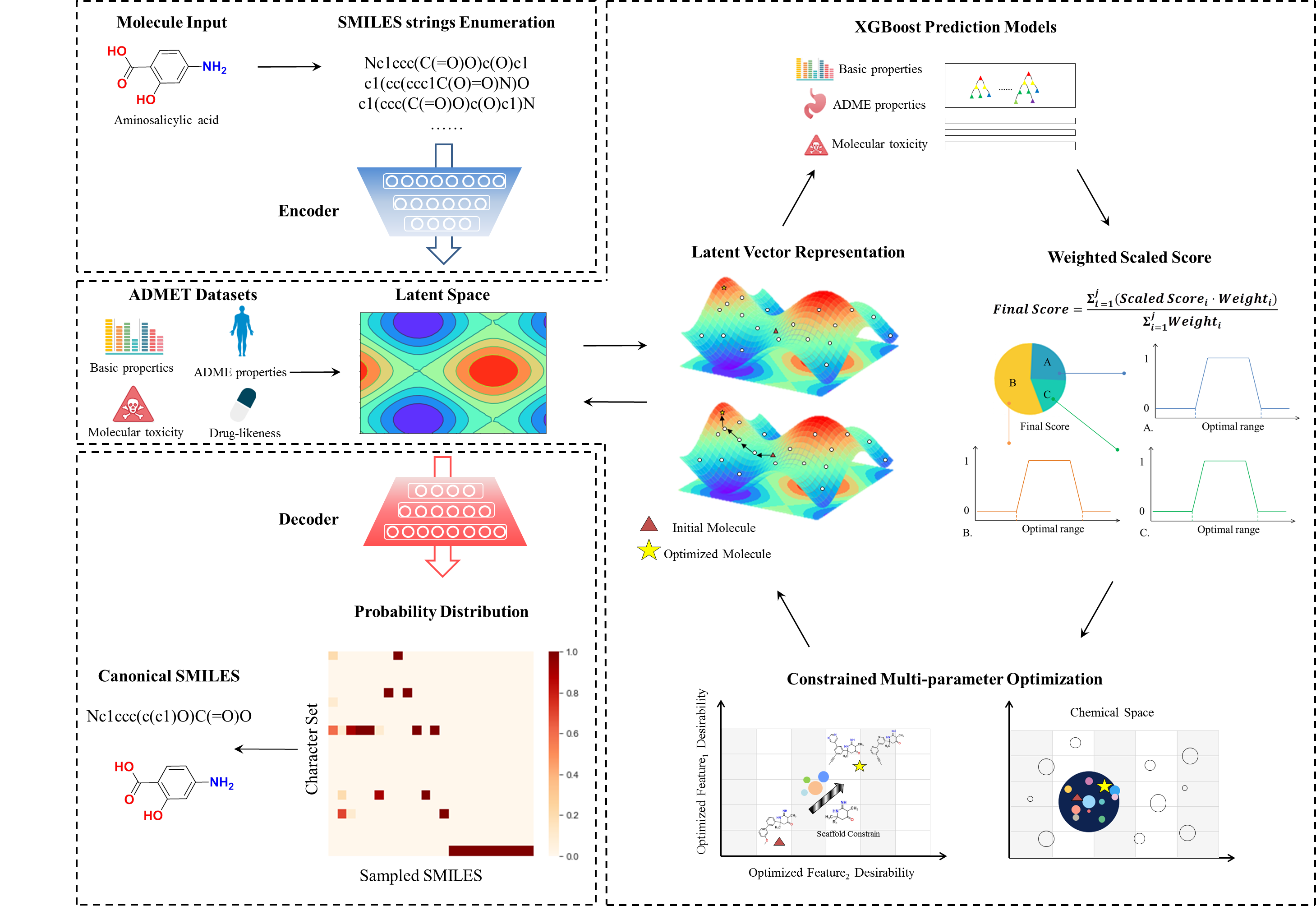
The SMILES Encoder allows users to transform a SMILES string to a 512-dimensional vector through the well-trained encoder network based on the Neural Machine Translation model. Owing to its consecutive, reversible, and informative features, such representation is recommended to define the chemical space of molecules.
The Descriptor Decoder can back-engineer the 512-dimensional vector to the corresponding uniform canonical SMILES string through the well-trained decoder network based on the Neural Machine Translation model. Such function provides a steered solution for de novo molecular optimization, as the possibility output of the decoder can be sampled.
The Molecular Optimizer, which combines the neural machine translation model and multi-objective particle swarm optimization, is designed to optimize the ADMET properties of molecules based on credible ADMET prediction models and the customized substructural constraints, thus effectively improving the drug-likeness of leads without the loss of potency.
Highlights
-
01
Neural Machine Translation Model
The Neural Machine Translation Model trained based on 17 million enumerated SMILES strings is able to capture the latent information behind molecular structures.
-
02
Reversible Machine Representation Learning
The consecutive, reversible and informative features of the calculated molecular representation make it possible to accomplish de novo molecular design.
-
03
Multi-parameter Optimization with Necessary Constraints
Directional optimization of ADMET properties with necessary substructure constraints ensures the balance between drug-likeness improvement and potency preservation.